Hosted Jupyter Notebooks and VS Code on Kubernetes
Jupyter notebooks are a powerful and popular tool that provides an interactive computing environment, combining code, data visualization, and explanatory text, making it easier to work with data and share insights. Data scientists use Jupyter notebooks for various tasks throughout the data analysis and machine learning lifecycle like exploratory data analysis (EDA), data preprocessing, visualization, model development, evaluation and validation, etc. For many of these usecases, just installing Jupyter Notebook on your laptop is enough to get started. However, for many companies and organizations, this is not an option and we need hosted Jupyter notebooks.
Why do we need hosted Jupyter notebooks?
- Access to resources: A lot of Data-Science workloads require heavy computing which cannot be supported by local machines of DS or MLEs. The hosted Jupyter notebooks can provide DS/MLEs access to powerful machines and GPUs.
- Data Access and Security: In many enterprises, DS/MLEs work on projects involving sensitive data which cannot be downloaded on the employee's laptop. So with hosted Jupyter notebooks, companies can provide access to DS/MLEs to work on sensitive projects.
- Reproducibility and Collaboration: Hosted Jupyter Notebooks are an excellent way to share executable code and results within a team. Since the development environment is the same, the results are 100% reproducible and team members can collaborate on projects.
How can a company provide hosted Notebooks to its data scientists?
Here are the options that a company can have today to provide access to Jupyter Notebook to its engineers:
Provision a VM for every Data scientist:
DS/MLEs can set up the environment and run a jupyter-server on a VM which can be used for running the workloads. Here is a simple guide on how you can run jupyterlab on an ec2 instance.
Use a Managed Solution:
Another option can be using a managed solution like AWS Sagemaker, Vertex AI Notebooks, or Azure ML Notebooks. While each of these methods has advantages, here are a few pros and cons of these in general.
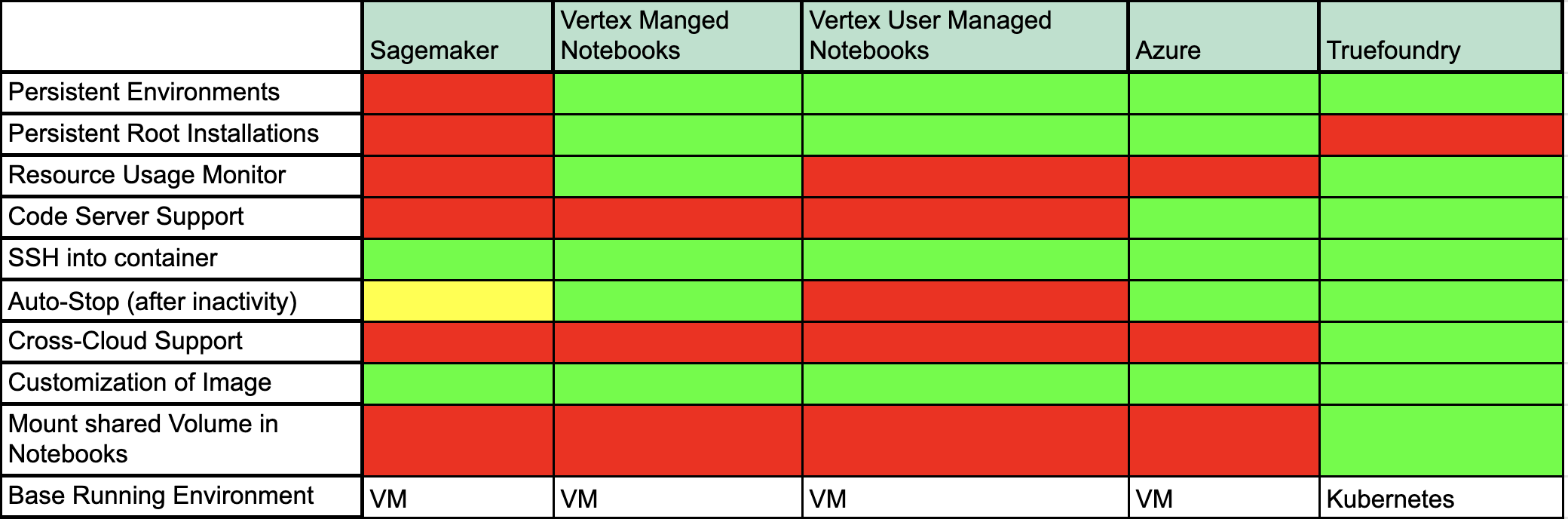
Let us discuss what each of these fields means:
- Persistent Environments: This refers to the persistence of the python environment. (Whether or not pip package installs and user-created environments are saved across Notebook Restarts)
- Persistent Root Installations: When root access is provided to the user, whether or not root package installations (e.g. apt install pkgs) are persisted across Notebook Restarts.
- Resource Usage Monitor: Whether or not the user can monitor CPU/Memory Usage from the notebook inside the notebook screen itself.
- Code Server Support: If the notebook instance can be connected to a hosted VS Code Server.
- SSH into the container: Ability to SSH into a running notebook and connect from the local machine
- Auto-Stop: Feature to stop the notebook after "Inactivity Period". Vertex Provides this feature in its "Managed Notebooks" version. Sagemaker has a way to stop notebooks after a time of inactivity, but the person needs to configure an "Init Script" for it instead of a simple deployment option which creates a bit of friction for the user.
- Cross-Cloud Support: This feature is exclusive to Truefoundry as you can run the notebook on any of the three cloud providers with the exact same user experience.
- Customization of Image: Starting the notebook with a particular set of packages pre-installed.
- Mount shared Volume in Notebook: Notebooks provided by each of the cloud providers (AWS/Azure/Vertex) allow creating a separate volume for each notebook but don't provide a way to mount a shared volume. For example, consider that an enterprise has a dataset of 500GB that is being used by multiple Data Scientists for different use cases. Now every DS needs to duplicate this data and pay for the cost of multiple volumes even when the notebooks are not running. This could potentially drain 100s and 1000s of dollars in cloud costs!
With Truefoundry you can mount a NFS type volume in read-only mode to multiple notebooks and thereby saving data-duplication costs!
Host Notebooks over Kubernetes:
Another option can be hosting notebooks over Kubernetes, but it comes with its own set of challenges as data scientists cannot directly interact with Kubernetes and need software in between that provides a simple interface to launch Jupyter notebooks. Lets us see what are the options available in this:
Kubeflow Notebook Operator:
Kubeflow helps to make deployments of machine learning (ML) workflows on Kubernetes simple, portable, and scalable. It has a notebook feature that helps to manage and run notebooks easily.
While Kubeflow is a large open-sourced project which provides a lot of features for Machine Learning use cases, it is very difficult to install and manage Kubeflow by yourself.
Host JupyterHub on Kubernetes:
JupyterHub is a great setup for multi-user use cases which helps in the optimal usage of resources. Deploying JupyterHub on Kubernetes can be done with an Open-Sourced project called Zero to JupyterHub with Kubernetes:
While there are a lot of solutions available right now, each solution comes with its own set of limitations. At Truefoundry we have tried to bridge this gap and tried to build a notebook solution that satisfies all the needs of a DS and also keeps costs in check. In the next section, we will describe our approach to building the notebook solution and the challenges we faced in making the same.
Truefoundry's approach towards building a Notebook solution
Truefoundry is a developer platform for ML teams that helps in deploying Models, Services, Jobs, and now Notebooks on Kubernetes. You can read more about what we do here. Our motivation for building a notebook solution was to simply enable experimentation and development on our platform. After studying all the solutions available, we decided to solve the pain points and missing features in the other platforms so that data scientists can have the best experience without incurring a lot of costs. A few things we wanted to enable are:
- Persist environments
- The ability for the user to install root dependencies dynamically and not be limited to a few sets of dependencies.
- Ability to run on the spot in certain cases since their pricing can be drastically lower. The experience can be bad in a few cases since the VM can be reclaimed, but we have found this to be quite rare in practice and the cost-benefit justifies the occasional hiccup.
- Ability to configure auto stopping to save costs.
Build on top of Kubeflow Notebooks
Kubeflow supports running notebooks on Kubernetes. It provides a number of features on notebooks out of the box. However, we wanted to address the issues we highlighted above in Kubeflow Notebooks and provide a seamless experience to datascientists and developers.
So, we had to make changes in the notebook controller, integrate it with Truefoundry's backend, and surfaced the notebooks on our UI.
We installed the notebook controller but bumped into some issues, because of which we had to make changes in the kubeflow-notebook-controller:
- Culling (Auto-Stop feature for Notebooks) doesn't work with custom endpoints. Kubeflow Notebook Controller relied on certain endpoint formats for culling to work. This limits the user to define the endpoint of their choice.
- Same Cull-Timeout across the cluster. This means that you can only set an "Inactivity Timeout" for notebooks on a cluster level. You cannot set different cull-timeout values for different notebooks.
- The default Python environment is not persistent
We solved the above two problems and launched the tfy-notebook-controller
and published it as a helm-chart Truefoundry's Public Charts repository. You can find the chart here.
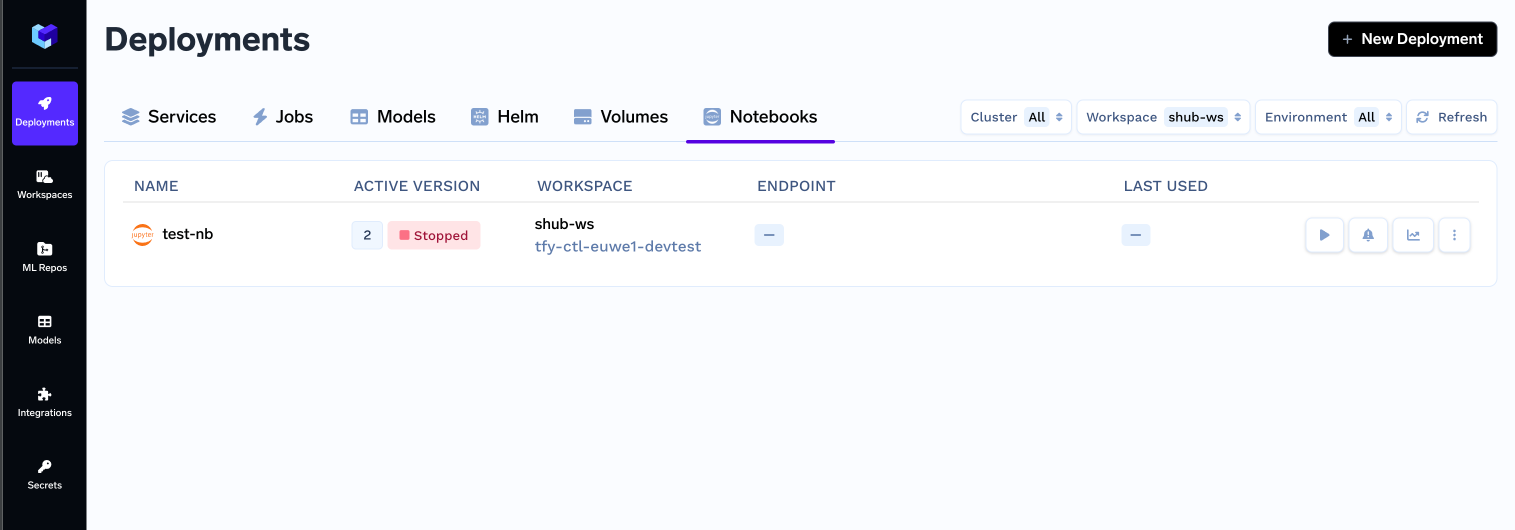
UI for creating, starting and stopping notebooks:
We created an easy-to-understand UI for data scientists to start notebooks. The user can customize the Idle Timeout (time of inactivity after which the notebook will be stopped), Persistent Volume Size (Size of Disk which stores the Dataset and code files), Resources (CPU, Memory and GPU requirements) and spin up the notebook!
With all these changes, we launched the v0 of our notebooks.
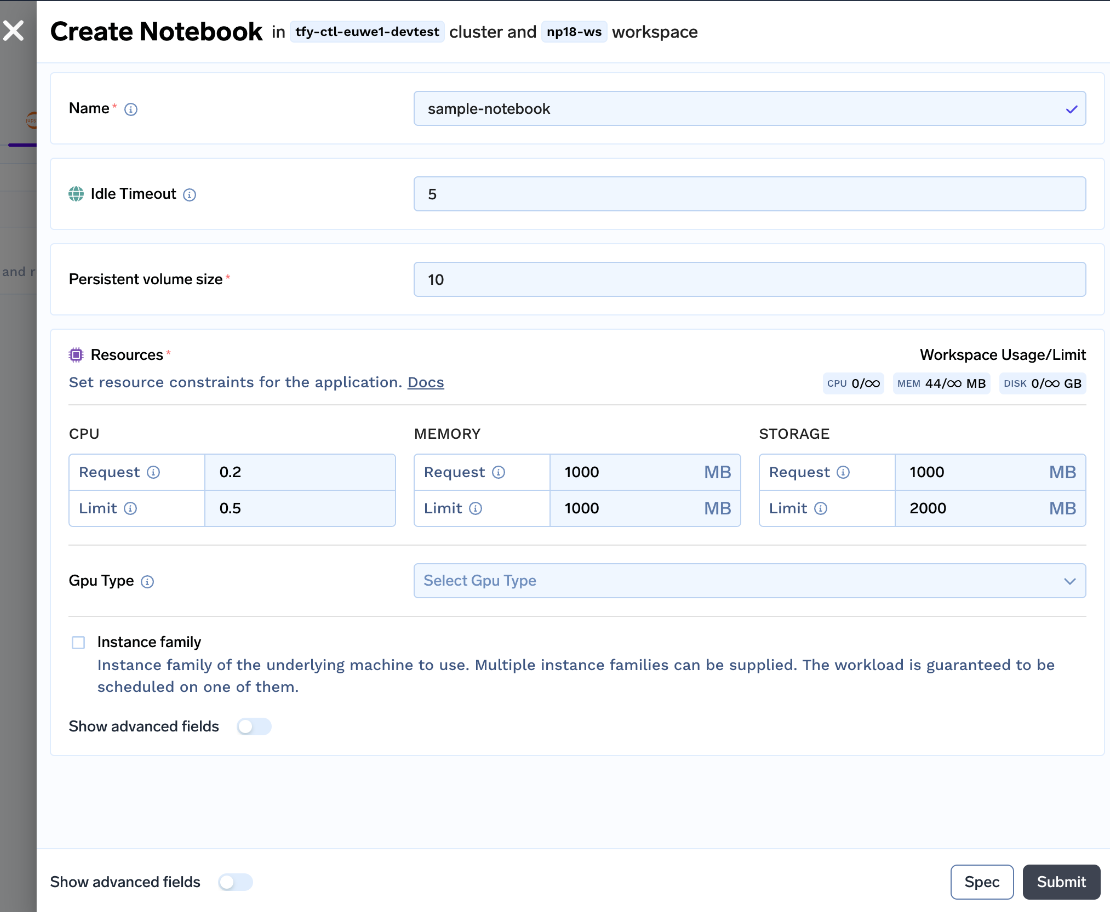
But still, we are a long way from a good user experience, lets us see the pros and cons of this approach:
Now these limitations are critical to solving as it blocks a lot of Data Scientist workflows which can be as simple as installing 'apt-packages' like ffmpeg.
Solving critical issues in user workflow
Till this point, we were using the pre-built images for Jupyterlab provided by Kubeflow. But since we need to solve the issue of non-persistent environments, allowing root access and installing apt-packages. We need to have our own set of docker images.
So let us look at how we solved these problems!
- Non-persistent Environments: The target was to make the base Python environment persistent so that users can easily use notebooks for their use cases.
jupyter-base- Add a .condarc file and set $HOME directory as the default environment path- Modify the .bashrc file to activate the jupyter-base environment by default- Install apt packages
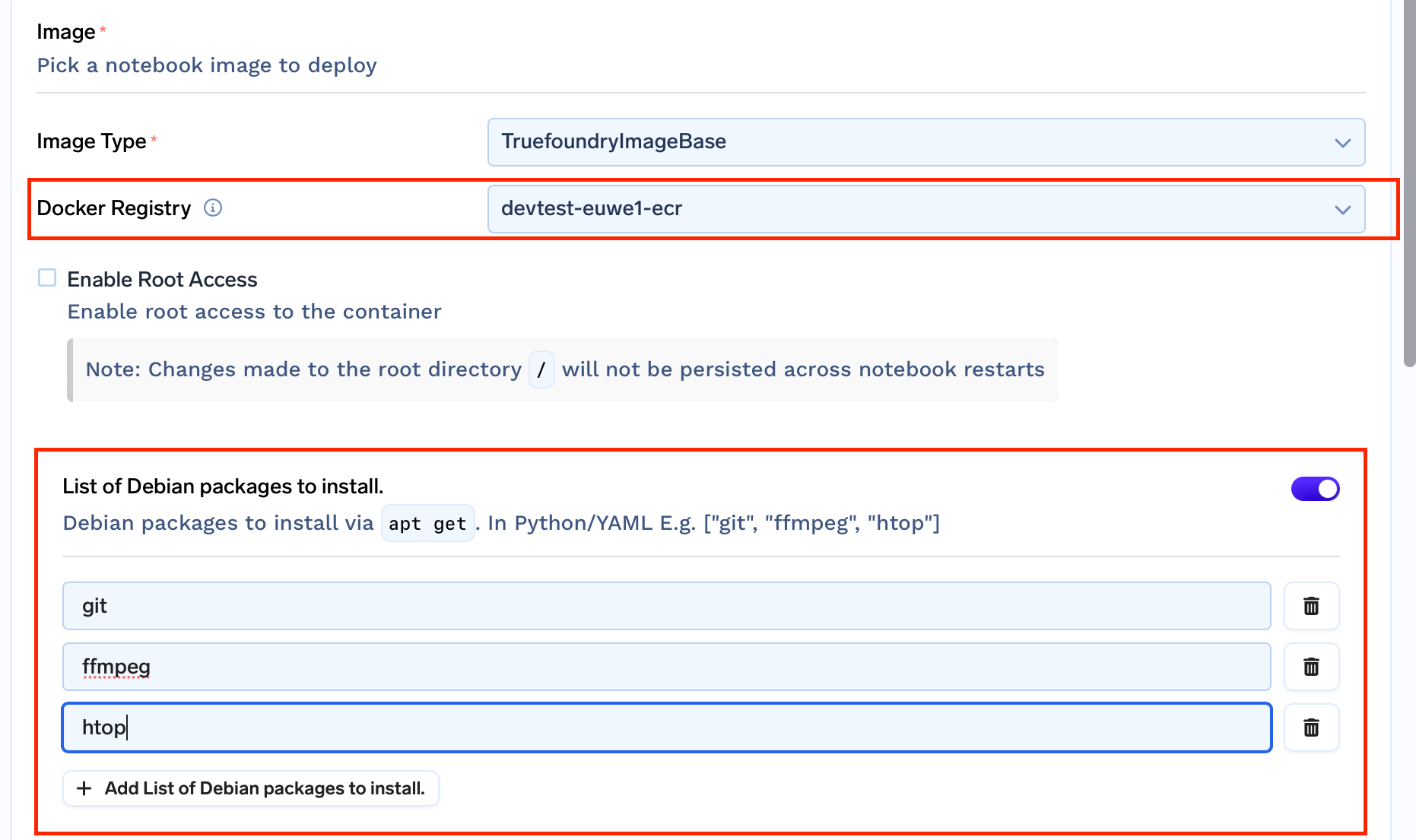
User has the option to provide a list of apt packages that they want pre-installed with the image.
We then add a build stage and create a custom image that comes with all the packages mentioned pre-installed in the notebook!
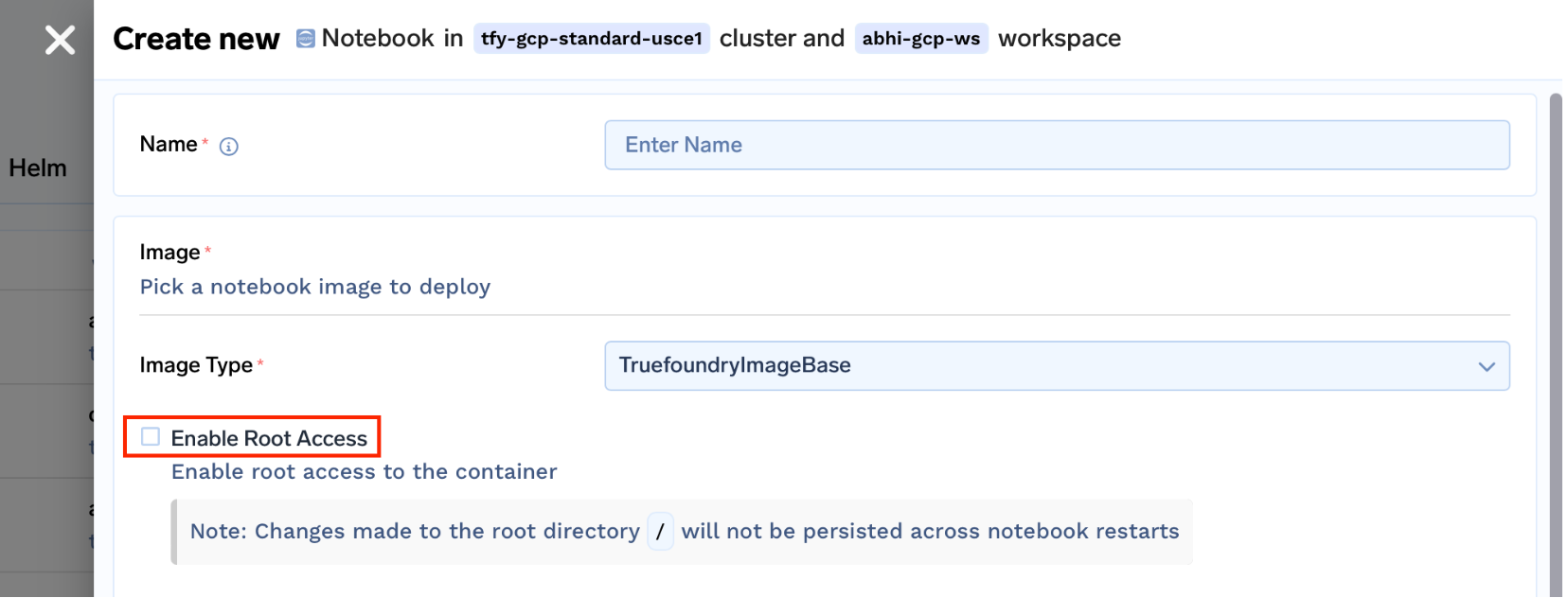
- Providing Root Access
In many cases, the user needs root access to install some packages and try out a few things quickly. For this, we created two images for each type of image.truefoundrycloud/jupyter:latestandtruefoundrycloud/jupyter:latest-sudo. Where the images with sudo provide n0-password sudo access to the user.
This was done by installing the sudo binary and adding the user to the sudoers list as described in this link.
Note: Since we are running notebooks on kubernetes with the home directory mounted, only the home directory will be persistent. The root packages installations will not be persistent across pod restarts. Please read this to get a better understanding of the same.
By solving these problems we solved most of the issues faced by a user and provide a decent notebooks experience. But with time we saw that users faced a few challenges which we will describe in the next section.
Usability issues in notebooks
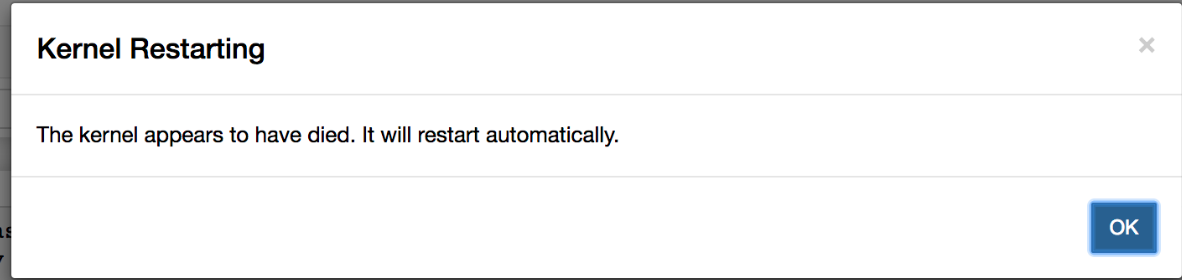
- Difficult to monitor the resource usage (CPU and memory for a notebook): Kernel Dies very often due to Out Of Memory issues. We often come across scenarios when we are running a notebook and the kernel dies due to one or the other reason, which can be tricky to debug.
- Modification of the Python environment can sometimes lead to a bad state where the notebook server fails to restart. This can happen due to multiple reasons like someone going and uninstalling
jupyterlabthe package. Since the environment is persisted, the notebook fails to start (once the current notebook is stopped) - Managing multiple environments is possible. But the user needs to manually add the conda environment to
kernelspecand ensure thatkernelspecis configured correctly which can issues.
Solving the Usability Issues
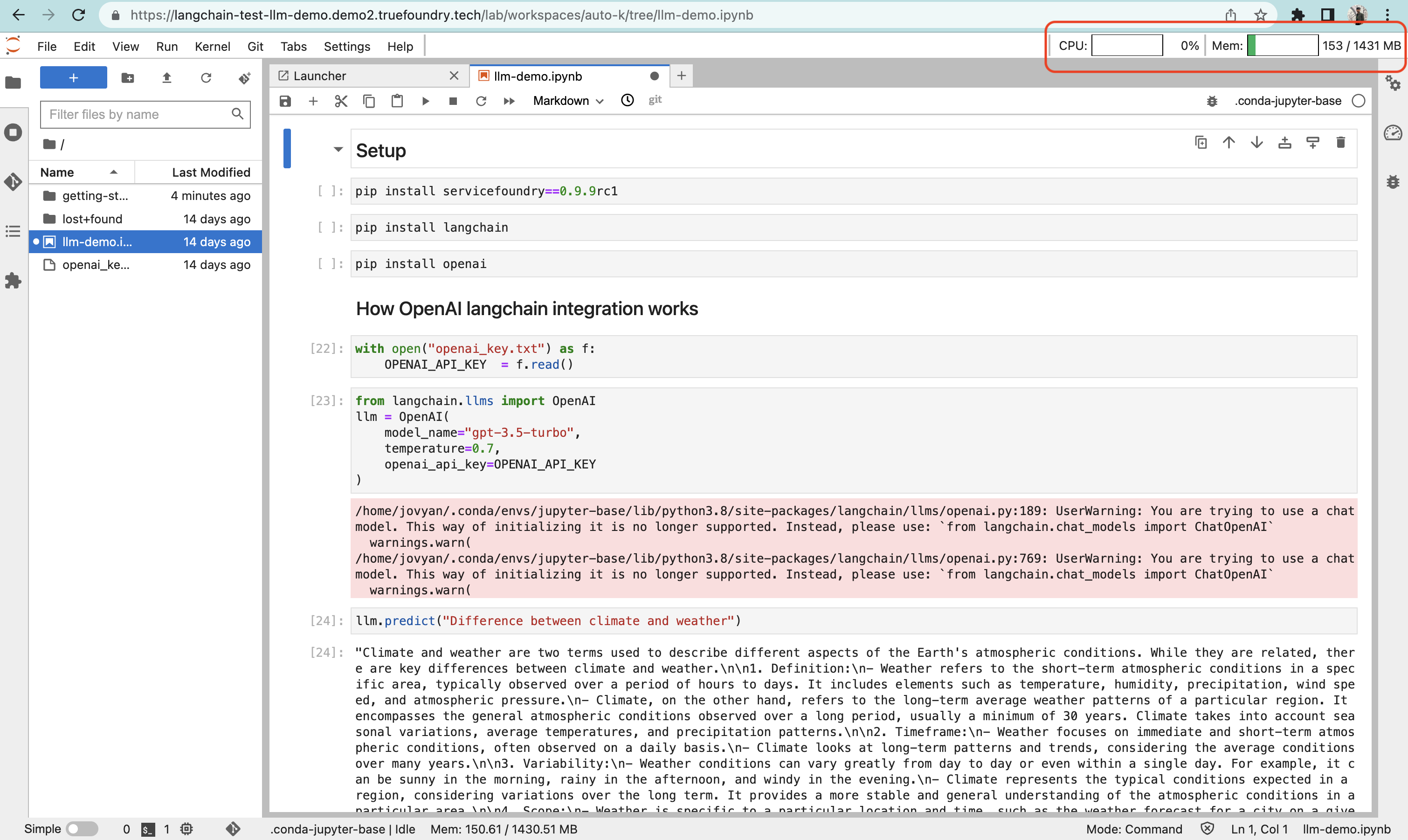
Adding resource usage metrics to the notebook:
We added the resource usage metrics to the notebook by installing the extension jupyterlab-system-monitor==0.8.0 and configured its settings in the init script by passing arguments while starting the Jupyterlab server.
...
jupyter lab \
...
--ResourceUseDisplay.mem_limit=${mem_limit} \
--ResourceUseDisplay.cpu_limit=${cpu_limit} \
--ResourceUseDisplay.track_cpu_percent=True \
--ResourceUseDisplay.mem_warning_threshold=0.8
This is what it looks like on the UI:
Separating out the kernel that runs the Jupyterlab server from the execution kernel
We need to ensure that whatever changes are done by the user in the home directory, the notebook should always restart without any issues. For this, we used the 'base' anaconda environment from /opt/conda the directory to start the Jupyterlab server.
Along with this, we created a separate environment in the $HOME directory, but this adds a kernel of the base conda environment to Kernels lists.
To solve for this we installed nb_conda_kernels to manage Jupyter kernels. We configured the init script to ensure that only the persistent Python environments show up in the kernel's list.
jupyter lab \
...
--CondaKernelSpecManager.conda_only=True \
--CondaKernelSpecManager.name_format={environment} \
--CondaKernelSpecManager.env_filter=/opt/conda/*"
With this, we get a guarantee that the notebook server will always start with whatever changes a user does inside the notebook.
It also eases out the management of multiple kernels. You simply need to create a new conda environment using the command conda create -n myenv and it starts to show up in the kernels list.
Adding Code-Server Support
While Jupyter notebooks solve a number of problems. There are a number of tasks at which it ceases to help:
- Development of service like a simple Gradio or Streamlit app. Users can write and run the code in Jupyter Notebook, but cannot view the the service in browser.
- If a user is working on a project which has code packaged into multiple files, the Jupyterlab environment is not suited for this development.
Considering these limitations, we decided to solve for the same. We added code-server support in order to provide a full IDE experience to the users in the browser.
By adding VS Code support, we enable users to do the following things:
- Develop and test services on your browser itself with the port-forwarding proxy of VS Code. This means your services deployed
localhost:8000can be made available at${NOTEBOOK_URL}/proxy/8000 - Easily manage code with proper packages, as this provides a full IDE experience.
- Make use of all famous VS Code extensions to boost productivity.
- Easily Debug applications by running them in debug mode and applying breakpoints.
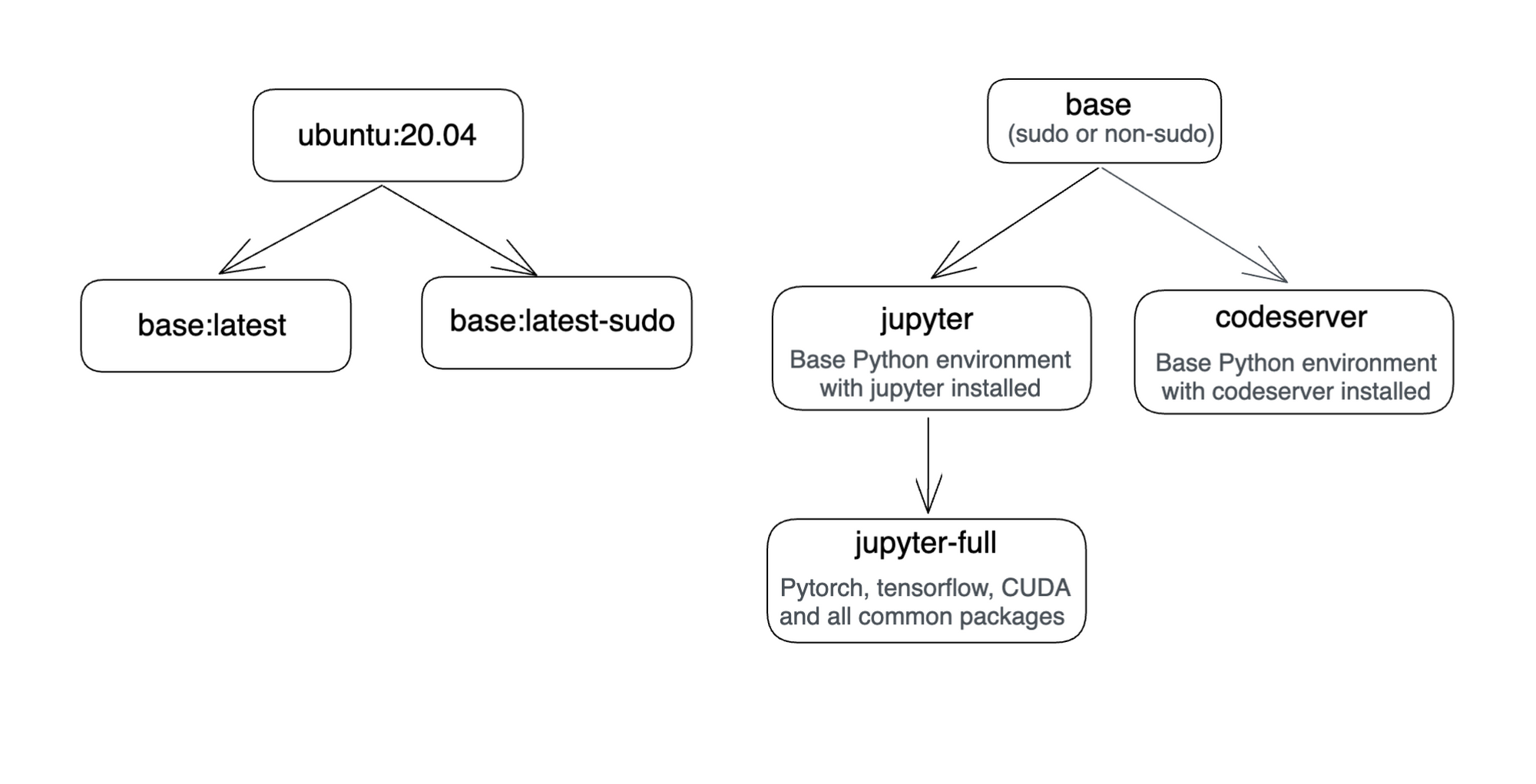
This was done by adding another docker image. Here is a diagram that shows Truefoundry's docker images.
SSH Access to your Notebook/VSCode:
While in most of the cases Hosted VS Code can solve the issue. But there can be cases (especially for Jupyter Notebooks) where the user gets stuck and needs direct access to the container running their Jupyter Notebook / VS Code Server.
So we have made that simplified by installing an ssh server in each of the notebooks and to connect to your container, you need to execute a simple command and enter your password:
ssh -p 2222 jovyan@test-notebook.ctl.truefoundry.tech

The power of this tool can be enhanced with your VS Code Extention called Remote Explorer where you can directly open all the files inside your VS Code!
Click here to read more about it
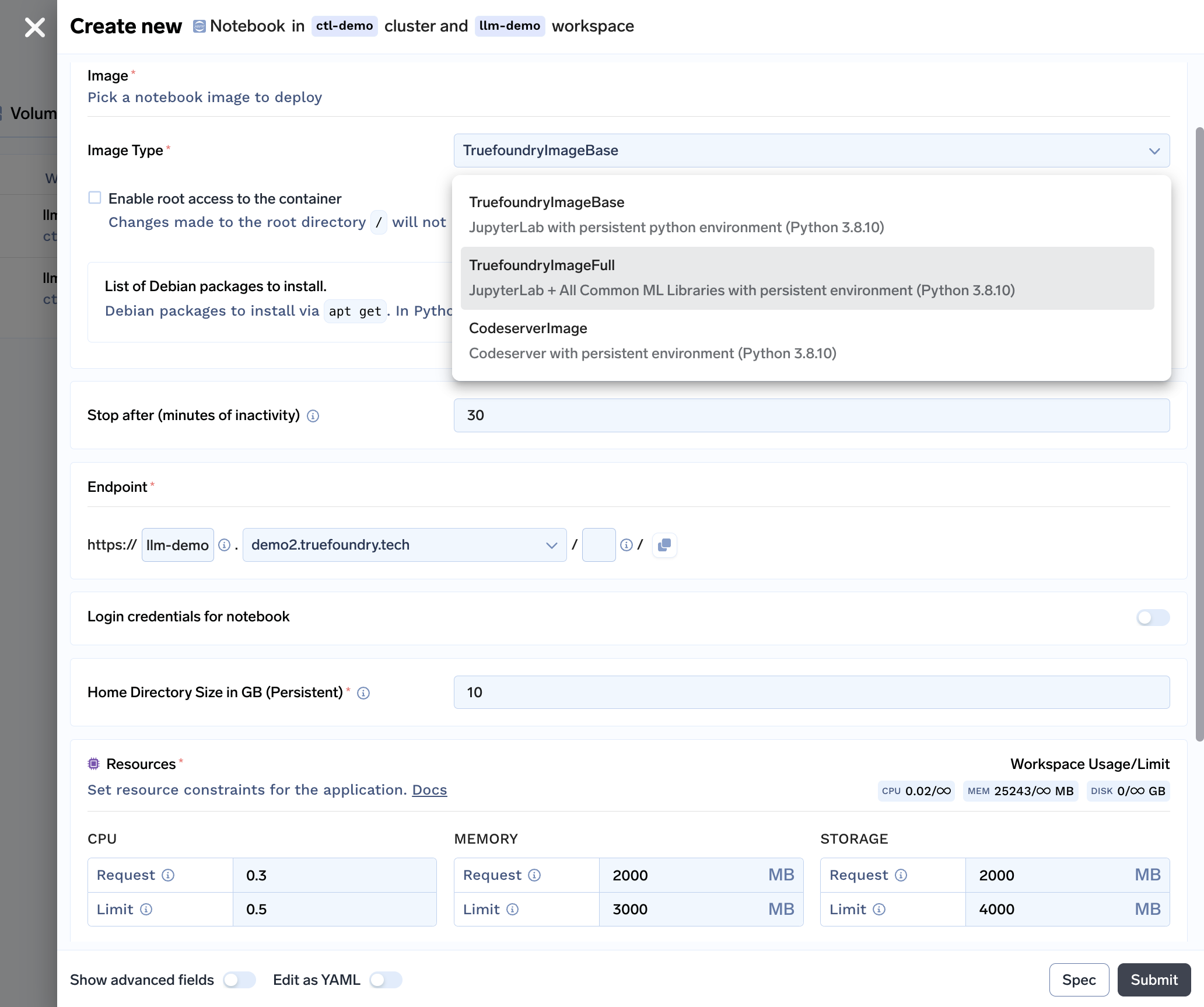
The Final User Experience:
With all the features bundled into our notebook solution, here is what our notebook deployment form looks like:
Price Comparison
Finally, let us compare the pricing of each of the managed solutions with Truefoundry.
Since Truefoundry works by deploying on the customer's cloud by connecting their Kubernetes Cluster, here is the pricing for Truefoundry running on different cloud providers.
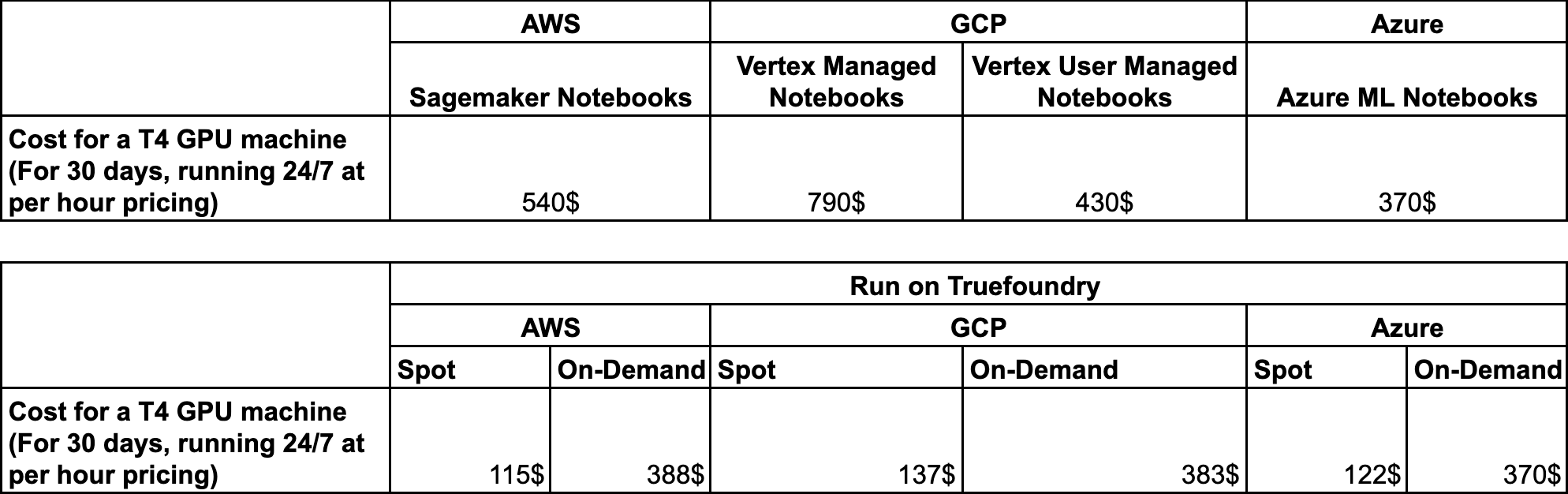
In the case of Truefoundry, you can actually save a lot of costs as:
- We do not charge any makeup cost over the top of the cost of Virtual Machines
- We support running notebooks on spot instances for non-Production workloads an addition of 50-60% more in cloud costs.
Conclusion
This was a brief about our effort in building the notebooks solution. You can join our Friends of Truefoundry Slack channel if you would like to discuss in depth about our approach or if you have some suggestions.
If you want to try out our platform you can register here!
